Element Bio and Google DeepVariant have teamed up to produce a benchmark for SNP and Indel variant calling using the CloudBreak Aviti short-reads sequencing method: https://www.biorxiv.org/content/10.1101/2023.08.11.553043v1
Google's software team assessed the accuracy of BWA+DeepVariant by comparing the results of the alignment and variant calling pipeline to the reference genome. They used a variety of metrics to assess accuracy, including:
Base accuracy: The percentage of bases that were called correctly.
Variant accuracy: The percentage of variants that were called correctly.
Indel accuracy: The percentage of indels that were called correctly.
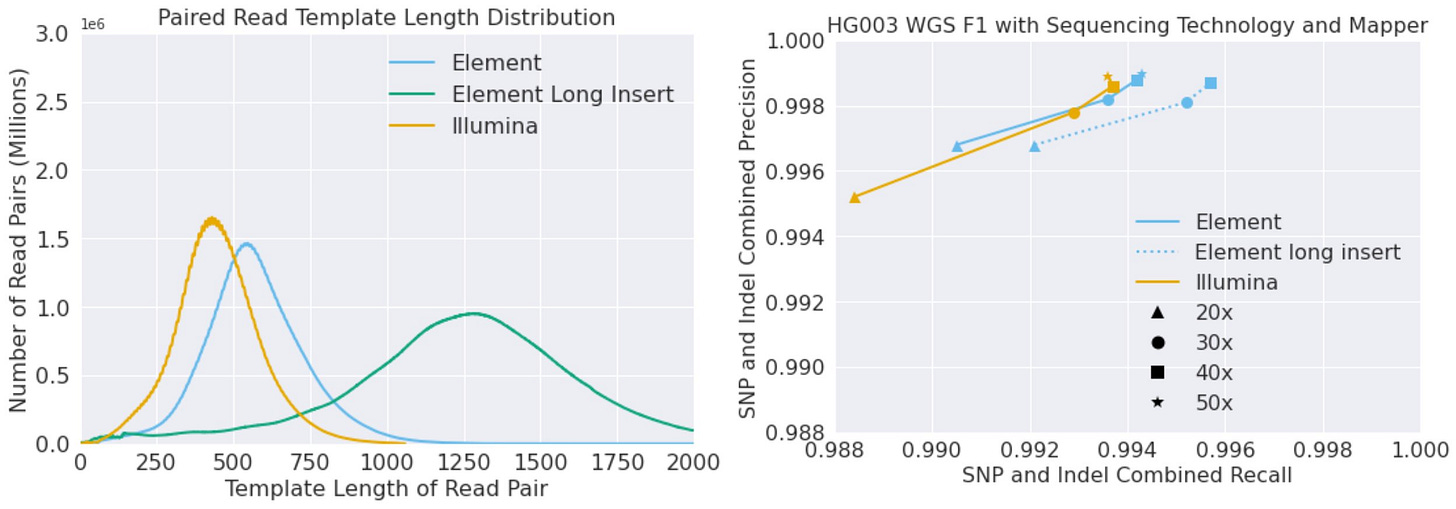
They found that BWA+DeepVariant v1.5 achieved higher accuracies with Element data than with Illumina data, especially at sample coverage between 20x and 30x. This is likely due to the fact that Element CloudBreak data is of higher quality than Illumina data.
To stratify to match coverage at sites, they divided the data into two groups: one group with coverage of 20x or higher, and one group with coverage of 30x or higher. They then compared the accuracy of BWA+DeepVariant on the two groups of data. They found that BWA+DeepVariant achieved higher accuracies on the group with coverage of 20x or higher, and even higher accuracies on the group with coverage of 30x or higher.
Overall, the results of Google's software team's assessment show that BWA+DeepVariant v1.5 is a highly accurate alignment and variant calling pipeline for both Illumina and Element data. However, it is especially accurate for Element data at high sample coverage.
Element data generates fewer candidates that need filtering because it has lower error rates in repeats and homopolymers. This is because Element data is generated using higher quality reads than Illumina data, which are less likely to be affected by these types of errors.
One way to benchmark the results of DeepVariant is to use the recent T2T assembly of ChrY to assess base-level concordance of reads with the reference genome. The T2T assembly was created using long-read sequencing technologies, such as those from Oxford Nanopore Sequencing. These technologies are able to generate reads that are much longer than Illumina reads, and they are less prone to errors in repetitive regions of the genome.
DeepVariant was able to call variants with high accuracy on the T2T assembly of ChrY. The predicted base qualities of the reads were in good agreement with the empirical error rates. This shows that DeepVariant is able to accurately call variants, even in regions of the genome that are difficult to sequence with short-read technologies.
Insert Sizes: Element Bio vs Illumina
The insert length between read pairs is the distance between the two ends of a pair of reads. Longer insert lengths can be beneficial for WGS accuracy because they can help to map reads to regions of the genome that are difficult to reach with shorter reads. This is because longer reads can span over larger regions of the genome, which can help to anchor them to the reference genome.
Element Bio AVITI sequencing is able to generate longer insert lengths than Illumina SBS sequencing. This is because AVITI sequencing uses a different sequencing chemistry than SBS sequencing. AVITI sequencing uses a rolling circle amplification method that creates a “nanoball” of DNA which can be attached to a surface at high density. Given this approach, longer insert sizes can be generated than with Illumina SBS.
The authors of the study found that longer insert sizes increased recall in WGS data. This is likely because longer inserts were able to map to regions of the genome that were difficult to reach with shorter inserts. The authors also found that graph mappers might have the potential to benefit even further from longer insert lengths. This is because graph mappers are able to take advantage of the longer inserts to build more accurate maps of the genome.
In contrast, Illumina SBS sequencing has been limited to shorter insert lengths in order to make sequencing cheaper. This is because the well size of the patterned flowcells has been made smaller and denser over time. The smaller wells make it more difficult to amplify longer inserts, which is why Illumina SBS sequencing is typically limited to insert lengths of 750 bp or less.
Overall, the longer insert lengths of Element Bio AVITI sequencing can be beneficial for WGS accuracy. This is because longer reads can help to map to regions of the genome that are difficult to reach with shorter reads. Graph mappers might have the potential to benefit even further from longer insert lengths.
Here are some potential consequences of this study for Illumina’s ILMN 0.00%↑ market prospects…