Multiplex-GAM and chromatin interactions: a brighter future for Epigenetic Engineering?
Will Multiplex-GAM find insights overlooked by Hi-C?
Headlines
There is a new paper published in Nature Methods, which details the new technique, Multiplex-GAM, to assay chromatin interactions in a population of cell nuclei from the assayed sample.
https://www.nature.com/articles/s41592-023-01903-1
The researchers from the Berlin Institute of Medical Systems Biology at the Max Delbrück Center describe a new technique based on the previously developed Genome Architecture Mapping (GAM) they've dubbed multiplex-GAM, which allows investigators to examine new and complex DNA interactions. The authors say that multiplex GAM brings a new way of looking at DNA interactions which was not possible with the currently popular genomic analysis technique Hi-C.
From the Genomeweb article:
““
In GAM, scientists take hundreds of thin slices of nuclei, each from individual cells and extract DNA from them which is sequenced and studied. However, in Hi-C, chromatin is cut into pieces using enzymes and then glued together again in such a way that two-way DNA interactions are revealed upon sequencing, which offers a limited view. The researchers say the differences between the techniques can be imagined as differences one would see on a color TV versus a black and white one. "With a black-and-white TV, you can see the shapes, but everything looks grey," co-author Ana Pombo, head of the epigenetic regulation and chromatin architecture laboratory at the Max Delbrück Center, says in a statement. "But if you have a color TV and look at flowers, you realize that they are red, yellow, and white and we were unaware of it. Similarly, there's also information in the way the genome is folded in three-dimensions that we have not been aware of."
““
Background into chromatin interactions
TADs, or topologically associating domains, are large genomic regions that are held together by topologically constrained interactions between DNA loops. Essentially, TADs are genomic regions that exhibit preferential physical interactions within themselves while having limited interactions with regions outside of the domain. In other words, TADs represent relatively self-contained regions within the three-dimensional structure of the genome.
It is debatable who came up with the term TAD first: some claim they were first identified in 2009 by the group of John Stamatoyannopolous at Stanford University. Others claim the term "topologically associating domains" was coined by a group of scientists led by Victor G. Corces and Job Dekker in 2012. They used high-throughput chromosome conformation capture (Hi-C) techniques to investigate the spatial organization of the genome. Through their research, they observed that the genome is compartmentalized into discrete domains, which they then named "topologically associating domains" or TADs.
TADs have become a significant focus of study in genome research due to several reasons:
Functional Units: TADs often encompass genes and regulatory elements that work together to regulate gene expression. By understanding the boundaries and interactions within TADs, researchers can gain insights into the mechanisms underlying gene regulation.
Disease Associations: TADs have been found to be associated with various diseases, including cancer, neurodevelopmental disorders, and other genetic disorders. Alterations in TAD structure and interactions can impact gene expression patterns and contribute to disease development.
Evolutionary Conservation: TADs have been found to be conserved across species, suggesting their functional importance. Comparing TAD organization between different species can provide insights into the evolutionary changes and conservation of genomic architecture.
Genome Engineering: Understanding TADs and their interactions is crucial for genome engineering technologies such as CRISPR-Cas9. TAD boundaries can influence the efficiency and specificity of gene editing by limiting the spread of genetic modifications to neighboring regions.
One of the key aims of genome-wide 3D chromatin folding assays is the detection of TADs. These assays use a variety of techniques to measure the three-dimensional structure of the genome, such as Hi-C and chromosome conformation capture (3C). By identifying TADs, researchers can gain a better understanding of how the genome is organized and how this organization affects gene regulation.
TADs are important in genome research because they provide a way to understand how the genome is organized in three dimensions. This organization is thought to play a role in gene regulation, and by identifying TADs, researchers can gain a better understanding of how this regulation works.
The merits of this new method
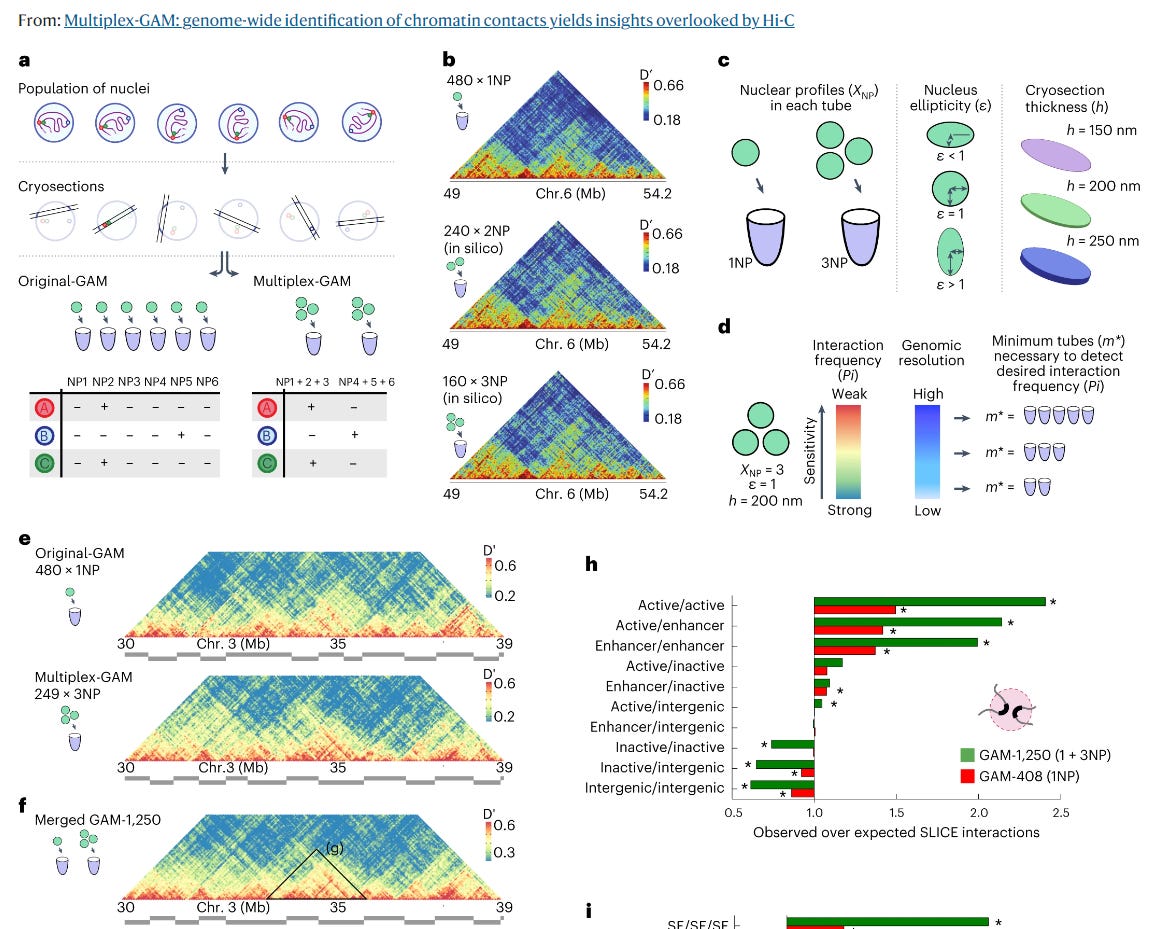
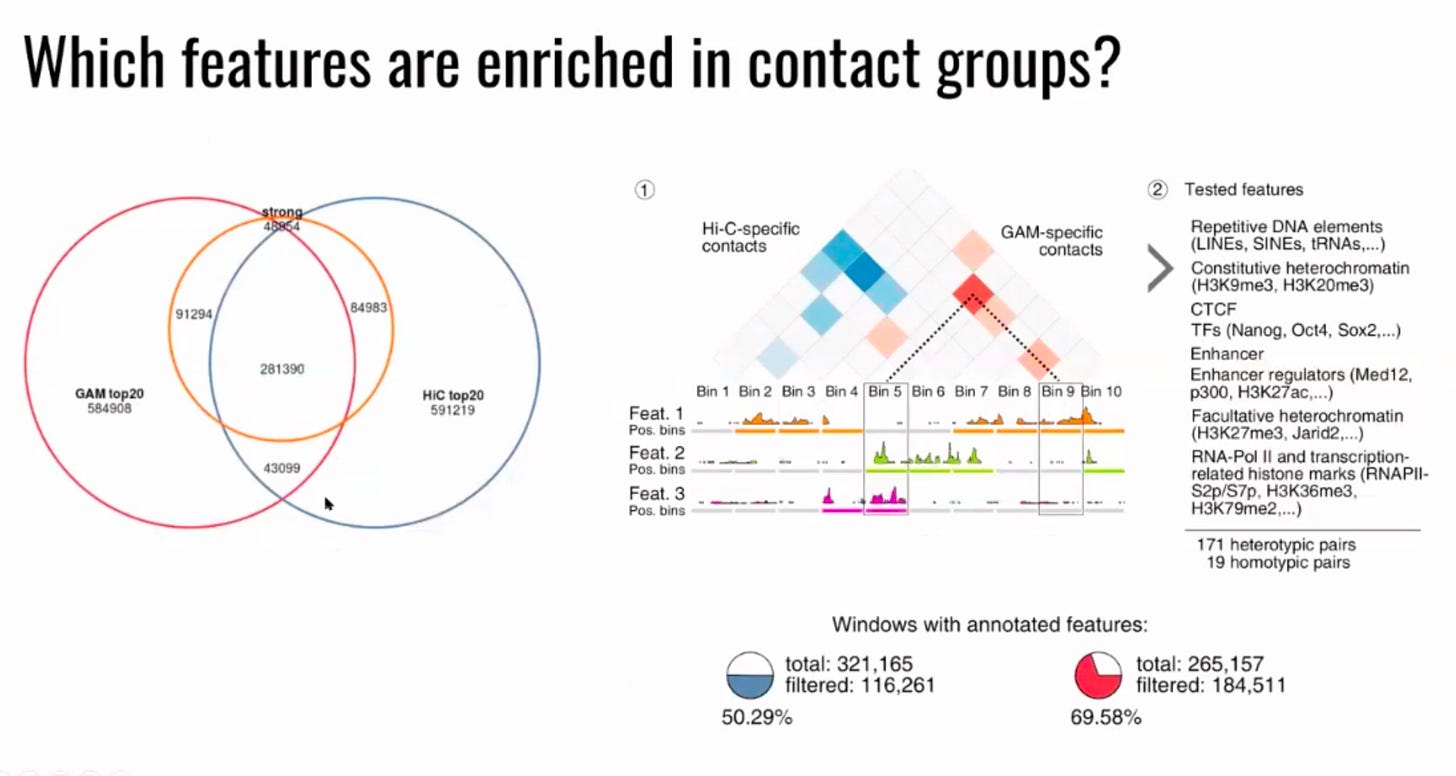
Here we analyse how much of a difference would the GAM method have over Hi-C.
They claim that: “GAM detects far more contacts at larger genomic distances than Hi-C, such as megabase-range contacts between super-enhancers, validated by single-cell fluorescence in situ hybridization (FISH) experiments”.
The previous version of Multiplex-GAM, called just GAM, was already a step ahead of HiC, capable of assaying chromatin interactions that were lost otherwise:
Epigenetic Engineering
The epigenome is the set of chemical modifications that regulate gene expression without changing the DNA sequence. These modifications can be inherited or acquired during life, and they can have a profound impact on how our genes are expressed.
Understanding how chromatin interactions at the genomic level regulate the epigenome is opening the door for new therapeutic approaches to treat a variety of conditions. For example, epigenetic engineering could be used to:
Silence genes that are associated with cancer.
Turn on genes that are associated with neurodegenerative diseases.
Promote the differentiation of stem cells into specific cell types.
There are several "epigenetic engineering" companies in biotech that are attempting to come up with therapies based on this approach.
If you want to read more about these, the information is at the other side of the curtain…