NGS and Multi-Omics November 2024 update
News from Illumina, Complete Genomics, PacBio, Element Bio and Singular Genomics
A busy month in Next-Generation Sequencing (NGS) and Multi-Omics. Several announcements by some of the key companies in this field, many at high profile conferences such as ASHG.
Complete Genomics (MGI Tech)
Complete Genomics (CG) is the U.S. subsidiary of MGI Tech in China, and specializes in next-generation sequencing (NGS) technologies, offering a comprehensive suite of products designed to meet diverse genomic research needs. Their product lineup includes high-throughput sequencing platforms such as the DNBSEQ series, which utilizes DNA nanoball technology to deliver high-quality data with rapid turnaround times. In the U.S. market, Complete Genomics has been expanding its presence, particularly since re-entering the market in 2023, in an attempt to avoid trade issues under the chinese MGI Tech brand. Despite this recent growth, Illumina continues to dominate the NGS landscape, holding over 90% of the market share in clinical genomics testing.
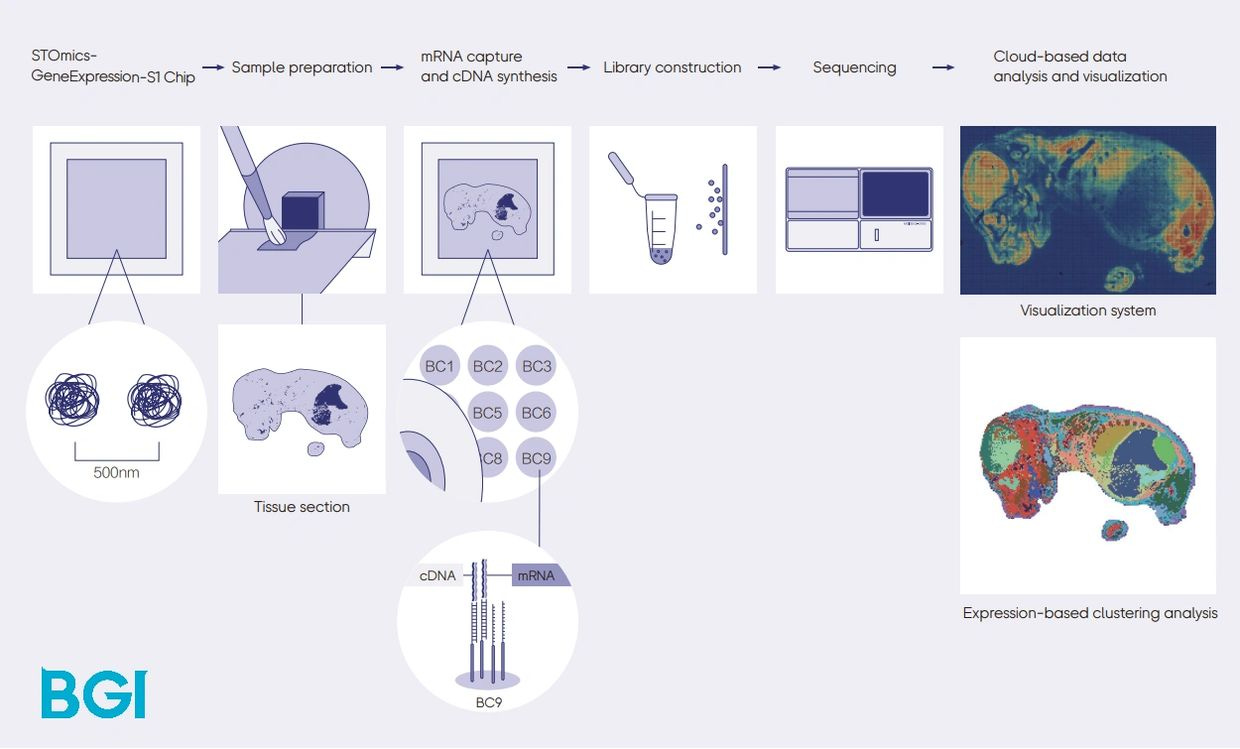
At the American Society of Human Genetics annual meetings (ASHG), Complete Genomics emphasized their recent work in multiomics and spatial biology. MGI Tech is already offering spatial omics products based on Stereo-seq. At ASHG, CG unveiled Go Spatial, an automated liquid handling solution for the Stereo-seq workflow, and FluoXpert, which makes the Stereo-seq products compatible with the mid-throughput DNBSEQ-G400 sequencers.
The type of tissue samples that can be processed with Stereo-seq started with fresh frozen, but a few months ago the technology expanded into FFPE tissue samples. There are two chip sizes for these, a 1cm x 1cm chip as well as a 2cm x 3cm chip, currently only for frozen samples.
CG wants to offer multi-omics solutions at an affordable price point, and their per-mm2 cost of Stereo-seq + NGS sequencing is in the $49-80 range depending on the chip size.
Illumina
Illumina also presented at ASHG, and the expectations were high, considering the noise that they made on social media in anticipation to the event.
Illumina MiSeq i100
The Illumina MiSeq i100 had already been announced in a previous event, and I did a deep dive on it in a separate post.
Illumina MiSeq i100 announcement
Illumina held an online event “Something big is coming. And it’s small”. The MiSeq i100 series, a slightly smaller in footprint than the NextSeq 2000, very similar in shape to a shrunk down NovaSeqX.
Constellation Reads (third attempt at long read technology)
Given that Illumina wasn’t allowed to acquire PacBio about a decade ago, the marketing power of Illumina has focused on short reads for the last 10 years, trying to convince customers that the combination of 150bp reads with clever ML in the DRAGEN software was enough for all applications.
But Illumina is making a third attempt at combining their proven short-read SBS technology with DNA fragment preps that would give the users the best of both worlds: high-quality short-reads turning into readouts of long fragments of contiguous sequences.
The first attempt was via the acquisition of Moleculo in 2013: the technology was splitting the pool of long fragments into barcoded groups serially, to then sequence short 150bp reads in each pool, with a diluting effect to the point where one could be more or less sure that all short reads with a given barcode were part of a unique long fragment, and thus simplify this small assembly task in this “divide and conquer” approach. The method never got much attention from users and was quickly phased out.
The second attempt was the CLR method, which Illumina got via stealth acquisition of a company in Australia called Longas. It had similarities with the barcoding approach of Moleculo, but crucially it had a “noisy polymerase” step which would intentionally add “markers” at random spots of the long fragments, which would help resolve difficult regions in this local assembly task via a software-based sequence consensus approach. Again, like Moleculo, this approach didn’t pick up much momentum.
The third attempt was announced in ASHG as Constellation reads, and it involves seeding the modified flow cells with long DNA fragments, and letting them land in a multitude of the wells where the tagmentase enzyme would chop them into smaller fragments in individual wells.
After the tagmentation step on the surface is completed, then the usual bridge amplification takes place, and from then on, it all looks like Illumina SBS sequencing as we’ve known in for many years.
Because of the pattern that these long molecules leave behind when they land, one can reconstruct in-silico what the long molecule was, as long as one has a reference sequence to compare against. Product number one is ready for human genomes, but presumably this would work with any kind of reference where one knows enough about the long stretches of DNA that is trying to reconstruct.
So this need for a reference is probably the most limiting factor for this technology: it gives what in the field is called “phasing information”, which is to say, if a 10kb molecule has 2 SNVs in it, one will known how these two variants are linked in that single 10kb molecule, but it’s no substitute for assembly-free approaches with true long read technologies like Oxford Nanopore.
This technology has been iterated at Illumina for a long time. In fact, Illumina R&D had internal versions of the Constellation Technology for more than 10 years, but this is the first time where it’s been publicly released as a product. The first iteration is capable of doing 16 DNA samples per run on NovaSeqX flow cells, with a 24hr run time, and integrated DRAGEN analysis for the results.
It comes with a simple 10 minute prep step which then loads the sample into the NovaSeq X sequencer.
5-base sequencing and Protein Sample Prep
Two more products that Illumina gave a bit more info about at ASHG where their version of the 5-base sequencing methodology that gives not only the T,C,A, and G bases that are the starting blocks of any DNA sequence, but also a fifth base, which can be an epigenetic mark, such as 5mC (DNA methylation). For this as well with for the Protein Sample Prep, Illumina is offering an easy to use solution that doesn’t require wet lab scientists laboriously pipetting for hours on end.
For the Protein Prep solution, Illumina has partnered with TECAN to provide a liquid handling solution that is “low hands-on”. This TECAN is not cheap, at around $500k, and can’t be used for other tasks. The first product will be able to handle either serum or plasma samples and can do 192 samples per week.
This protein prep that Illumina has now described in some level of detail is in collaboration with SomaLogic, and will compete with the similar offering that Olink (now part of Thermo Fisher) has in the market.
PacBio Vega
There were lots of rumours about the new instrument that was going to come out of Pacific Biosciences, and some pointed to a higher throughput short-read sequencer, to compete with the NovaSeqX product line from Illumina, but most rumours were pointing in the direction of a smaller footprint version of the long-reads Revio instrument. The later turned out to be true, in the shape of a washing-machine sized Vega benchtop instrument.
This is a lower CAPEX version of the Revio ($169,000 vs $599,000) but with a price per gigabase higher than the Revio ($1,100 per run of 60Gb of data).
Together with the recent announcement of the improved SQPR chemistry, PacBio claims that now the price range of a 20-30x human genome goes from $500 for a 20x Revio run, to the ~$2,000 for a 30x (90-120Gb) of the Vega product accounting for amortization of the machines. The full table of specs can be seen at http://bit.ly/ngsspecs
Singular Genomics
Singular Genomics OMIC 0.00%↑ had announced months ago their first foray into NGS + Multi-Omics (NGSX) with their G4X instrument. The company has recently given more details on what this product looks like: it is capable of imaging 40cm2 of a biopsy, as a combination of In-situ transcriptomics, proteomics and fH&E, with subcellular resolution.
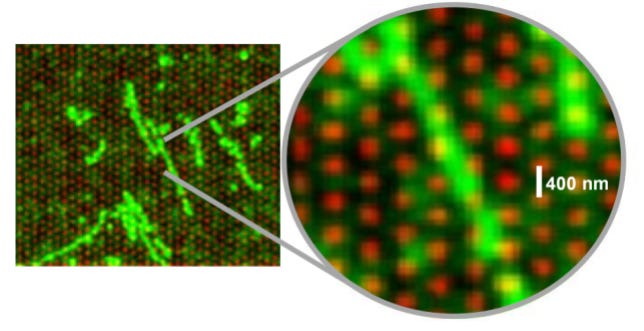
The transcript counting happens via the design of probes in the shape of padlocks, which are then amplified to form the same type of “nanoballs” as with the protein labelling, which happens with antibodies conjugated with DNA barcodes as shown below.
Since this is an NGS machine, it’s able to tell where these probes/barcodes are located within the sample, and it does so with subcellular resolution.
The key to all these multi-modal datasets is good software to find the patterns in the data. Singular Genomics shows how using unsupervised multi-modal clustering one can perform differential gene expression analysis and cell type classification where the RNA + Protein Cell type matches very well what the traditional H&E data shows.
Element Bio
Element Bio had an event last month showcasing similar NGSX capabilities for their AVITI24 instrument and Teton reagents. The main difference between the two is that Singular is able to process FFPE tissue biopsies, whereas the first iteration of Element Teto is for disaggregated live cells that will land on the flow cell for cytoprofiling. A deep dive of the Element Bio announcements is available on the blog post below.
Next-Generation Sequencing October 2024
We’ve had a busy month in Next-Generation Sequencing (NGS) and Multi-Omics (NGSX), with announcements from Element Bio, Illumina , Ultima Genomics and GeneMind as highlights.
Strategic Positioning
Multi-omics applications, with Spatial at the forefront of growth, are rapidly expanding. This is not only for basic research, but also more and more for translational research, clinical trials, and even clinical diagnostics. Because of the history of NGS, the field has been heavily biased towards genotyping (including Whole-Genome Sequencing WGS). Now with the kind of spatial omics and spatial imaging technologies that are becoming more and more available, spatial phenotyping will increasingly gain adoption. This is the “pheno” complementing the “geno” of the last two decades.