Single Cell Omics, Spatial Omics and Spatial In Situ Imaging
Current landscape and hypothetical plays in the near future
Main modes and players
In recent years, methods for encapsulating individual cells and analysing their DNA, RNA or protein contents have become more established, with commercial support from Biotech and Life Science Tool providers similar to what Next-Generation Sequencing received over the past decade or so. There is now a growing effort to transform academic lab-developed tools for Spatial Biology, especially those that use NGS read-out, into commercial products that are robust and can support a range of assays, such as protein and gene panels, and different biopsy configurations.
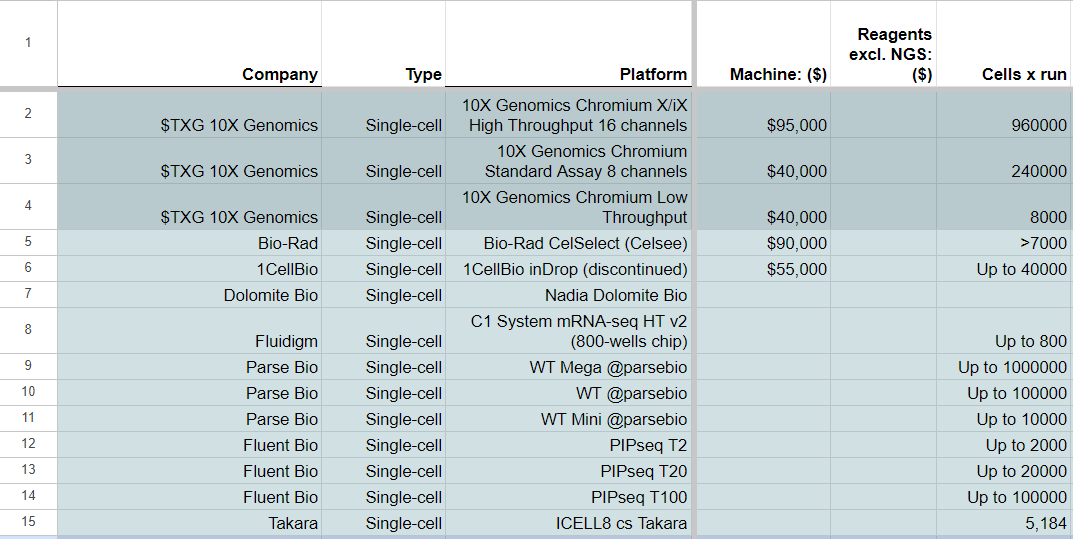
In the realm of single-cell omics and spatialomics, two companies, 10X Genomics and NanoString, have initiated a competition for market domination, both 10X Genomics and NanoString kicked off the race for market domination with their Chromium and Visium line of products, and GeoMx respectively. Other entrants such as Parse Bio and Fluent Bio have come up with instrument-free versions of the single-cell transcriptomics methods, with cells per run ranges similar if not higher than the two established players.
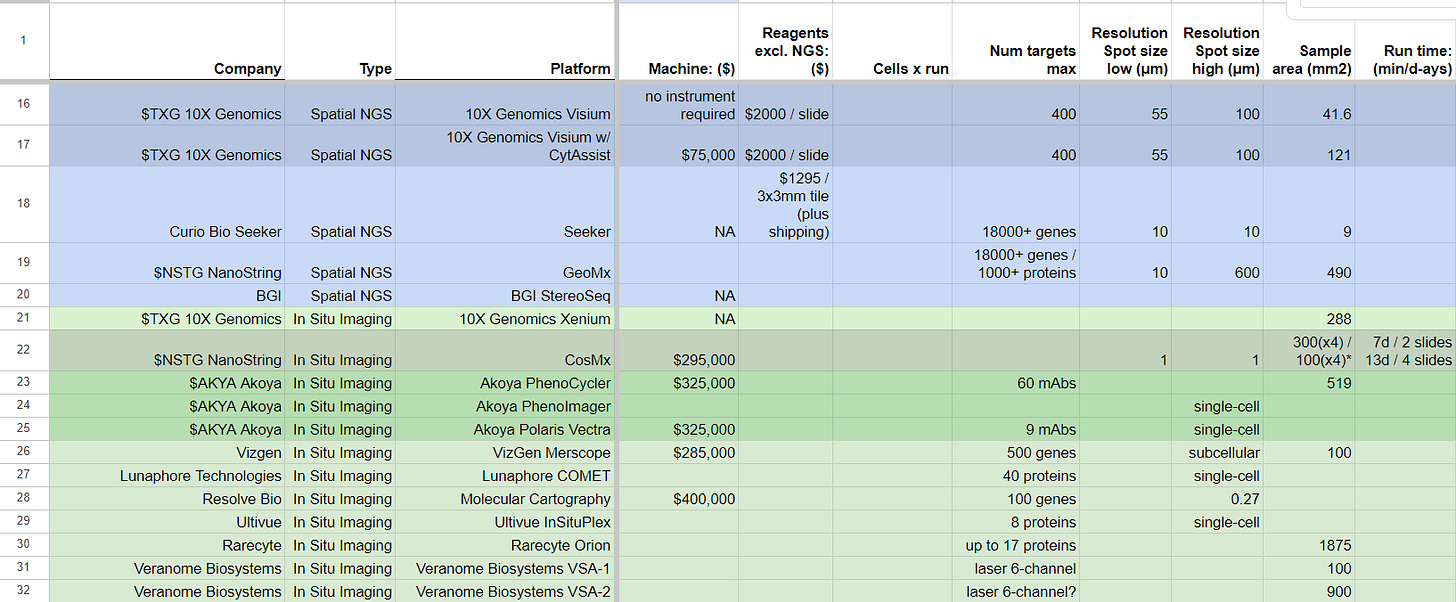
The same two companies, 10X Genomics and NanoString, have anticipated the value of imaging-based equivalents of their spatialomics products for assessing biopsies, and have come up with their Xenium and CosMx product lines, respectively. Other companies, including Akoya Bio, Vizgen, Lunaphore, Resolve Bio, Ultivue, Rarecyte, and Veranome Bio, have also entered the market with their own alternatives to Xenium and CosMx. These companies have backgrounds in traditional high-resolution microscopy and have partnered with antibody producers or developed their RNA-based probes to create multiplex panels of binding/hybridization plus imaging assays.
Specifications such as resolution (spot size or imaging), number of targets, and sample area are now determining which companies are leading the way in this emerging field of Spatial Biology.
Applications
The current segments of applications for Single-Cell, Spatialomics and Spatial In Situ Imaging can be described as:
Life Sciences: academic-based customers, with large atlas projects and a long tail of smaller research customers.
Applied biotech: privately/VC funded biotech companies using these tools for their discovery programs or internal diagnostics.
Diagnostics: The field of diagnostics is currently underdeveloped and has not yet reached its full potential. However, it is expected to experience rapid growth in the coming years, particularly with the development of Spatial Biology tools. These tools have the potential to revolutionize the way we diagnose diseases, and this is especially true for pathology diagnosis.
Currently, trained pathologists perform diagnosis using H&E/IHC-based pathology, by examining biopsy samples under the microscope. However, with the advent of Spatial Biology tools and the increasing use of machine learning and artificial intelligence, we can expect to see a shift towards integrated diagnosis in the future. This means that biopsy samples will be turned into pathology reports using these advanced technologies, with the results then being confirmed by a trained pathologist.
By leveraging the power of Spatial Biology tools, machine learning, and artificial intelligence, we can create more accurate and reliable diagnoses. This has the potential to greatly improve patient outcomes and provide a more personalized approach to healthcare. While there is still much work to be done in this field, the future looks bright for the development of innovative diagnostic tools and techniques.Most of these are based on either the full transcriptome assaying of either single cells or biopsies, or equivalents in panels of proteins (bound by fluorescently-conjugated antibodies) or RNA probes.
Single cell Methods
In recent years, there have been various technologies developed for single-cell analysis. Single-cell transcriptome methods continue to be the most common and best commercially-supported, with the full poly-A transcriptome (scmRNA-seq) being the most common, but there are others:
Single-cell mRNA sequencing (scmRNA-seq): This approach isolates and sequences the mRNA molecules from individual cells, allowing for high-throughput transcriptome analysis.
Single-cell TCR/BCR sequencing: This method enables the identification and quantification of T and B cell receptor sequences in individual cells to study the adaptive immune system.
CyTOF (Cytometry by Time-Of-Flight): This method combines mass cytometry with single-cell analysis to enable the detection of multiple cell surface markers in individual cells. It’s one of the now classical methods from 2011 sold by Fluidigm.
CITE-seq is a method of labeling cell-surface proteins with uniquely barcoded antibodies, which can then be captured and quantified along with the RNA molecules in single-cell RNA sequencing.
REAP-seq (RNA expression and protein sequencing assay): This method combines RNA and protein detection using oligonucleotide-labeled antibodies. The oligos are cleaved and barcoded, allowing for simultaneous detection of RNA and protein in the same reaction. There is a commercial version sold by Fluidigm.
ATAC-Seq (Assay for Transposase-Accessible Chromatin using sequencing), which has been used to understand chromatin accessibility in single cells. It uses a transposase enzyme to insert sequencing adapters into open chromatin regions in individual cells. The chromatin is then fragmented and sequenced to identify regions of accessible chromatin. While this technology has shown promise, it is not as prevalent as some of the other single-cell analysis tools. Other variations of the original method are scDual-ATAC-seq, sci-ATAC-seq, SHARE-seq, SNARE-seq, scTHS-seq, etc.
Perturb-seq: yet another technology that has gained attention for CRISPR perturbation screening. There have been many published examples of this technology, and it is also being widely applied in the private datasets of biotech and big pharma companies. CRISPR perturbation screening involves the use of CRISPR-Cas9 gene editing to perturb genes of interest in single cells, allowing researchers to identify gene function. Other variations of the original method are CROP-seq, CRISPR-UMI, Mosaic-seq, CRISPR-scATAC-seq, etc.
Single-cell WGA: although DNA sequencing of single-cell assays is being pushed by companies like Mission Bio, this technology has not gained as much popularity as some of the other tools mentioned above, although they have a niche in haematological cancers. However, there are still ongoing efforts to improve and expand the use of DNA sequencing in single-cell analysis. Variants of this method are DOP-PCR and MDA.
Overall, the field of single-cell analysis is constantly evolving, with new technologies and techniques being developed and refined. These advancements hold great promise for improving our understanding of biological systems and unlocking new insights into disease mechanisms.
Single cell vs Spatial
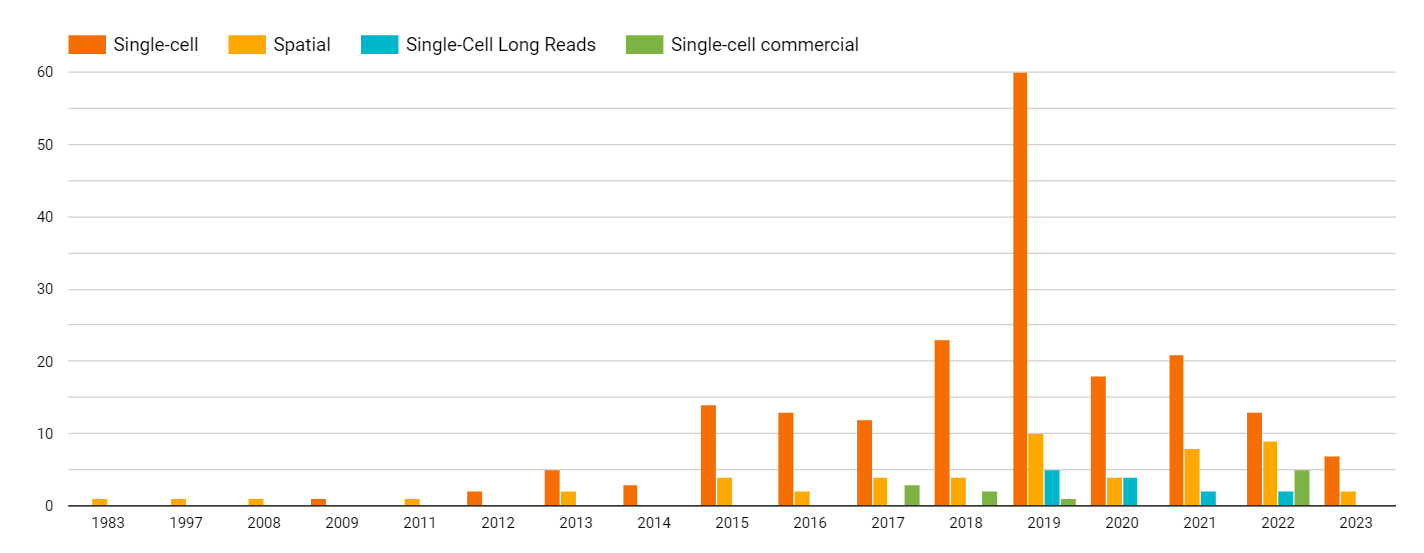
The field of single-cell analysis has been around for about 5 years longer than Spatial Biology approaches, and as a result, it has reached a greater level of maturity and wider adoption. This is reflected in the number of academic and commercial publications on single-cell methods, which peaked in 2019.
However, the trend is expected to shift towards Spatial Biology approaches in the coming years, with a projected peak in the number of publications around 2024 or later. Despite the larger amount of information contained in Spatial-omics or Spatial In Situ datasets compared to single-cell disaggregated samples, the familiarity of users with Spatial Biology approaches and their level of commitment to using these tools is still relatively low compared to single-cell methods. This is likely due to the more recent development of Spatial Biology tools and the need for more time for users to become familiar and comfortable with them.
As Spatial Biology tools continue to mature and become more widely adopted, they hold great promise for unlocking new insights into complex biological systems and disease mechanisms, and for improving diagnostics and treatment strategies.
Single-cell and Spatial methods
Unmet needs and clinical utility
Spatial-omics will only take over to the same extend as single-cell when we have products as robust as the Chromium or the nCounter NGS for the later, which hasn’t happened yet. The Visium HD product by 10X Genomics hasn’t been released yet, and it will require a $75,000 CytAssist instrument to run the assay on the biopsy slides. In comparison, the NanoString GeoMx is of a resolution much better to the instrument-free low resolution Visium by 10X Genomics, but most of the published GeoMx datasets still contain NanoString’s employees in their author list. This usually happens when a tool is still not robust enough for customers to consistently and regularly run on their own, without assistance from the specialized field engineers of the company offering such tools. The final two worth mentioning in Spatial-omics are:
Curio Bio, a newly emerged company commercializing slide-seq2, and promising similar specs as both Visium HD and GeoMx in an instrument-free solution.
BGI StereoSeq method, still not available commercially, which is promising a large sample area and high resolution with an NGS-readout linked to MGI Tech DNBSEQ sequencing.
On the Spatial In Situ Imaging front, both the Xenium and CosMx instruments are starting to land in customers’ benches, and together with Akoya Bio and Resolve Bio products, we see more of this happening.
It is likely that as Spatial Biology tools become more robust and user-friendly, they will become increasingly popular and widely adopted in the scientific community. This may lead to a shift in the balance between single-cell and Spatial Biology approaches, with the latter eventually becoming more prevalent. Additionally, as more and more datasets are generated using Spatial Biology techniques, the field of Machine Learning and Artificial Intelligence will likely play an increasingly important role in analysing and interpreting this data. This could eventually lead to a future where pathology reports are generated by machines rather than human pathologists, though this is likely to be many years away. Finally, as competition in the Spatial Biology market heats up, we can expect to see continued innovation and improvements in the technology, leading to even more powerful and versatile tools for researchers to use in their work.
More speculative comments and rumours below…