I will highlight below the announcement from Singular Genomics OMIC 0.00%↑ that coincided with day 1 of the AGBT24 meeting.
The AGBT meeting, formally known as the Advances in Genome Biology and Technology meeting, is a series of three major conferences focused on different aspects of genomics:
1. General Meeting: This is the flagship event, held annually in February. It's the preeminent conference for top researchers, leaders, and innovators in the field of genomics.
2. Agriculture Meeting: AGBT-AG. This new conference takes place in April and focuses on the application of genomics in agriculture.
3. Precision Health Meeting: Held in September, this conference brings together experts in genomics, healthcare, and industry to explore the future of precision medicine.
Singular Genomics is a life science technology company developing tools for Next-Generation Sequencing (NGS) and multiomics applications. They aim to empower researchers and clinicians by providing faster, more accurate, and flexible sequencing solutions.
Current Product:
G4 Sequencing Platform: This is their commercially available benchtop sequencer. It boasts several key features:
Speed: Delivers results in as little as 24 hours for some applications.
Accuracy: Utilizes novel chemistry and advanced engineering to minimize errors.
Flexibility: Accommodates diverse sample types and sequencing applications.
Scalability: Offers up to 4 flow cells for increased throughput.
User-friendly: Designed for both novice and experienced users.
The G4X announcement
The 10 minute presentation is very reminiscent of recent Apple product announcements. Drew Spaventa (CEO & Co-founder) gives an intro from the Singular Genomics HQ in San Diego.
Gives a timeline of the G4 instrument:
This is a mid-throughput instrument comparable to the Illumina NextSeq or the Element Bio AVITI instruments:
Makes the point on why Spatial Biology is difficult: scale, workflow, cost, and gives a timeline on what has happened inside OMIC in this area.
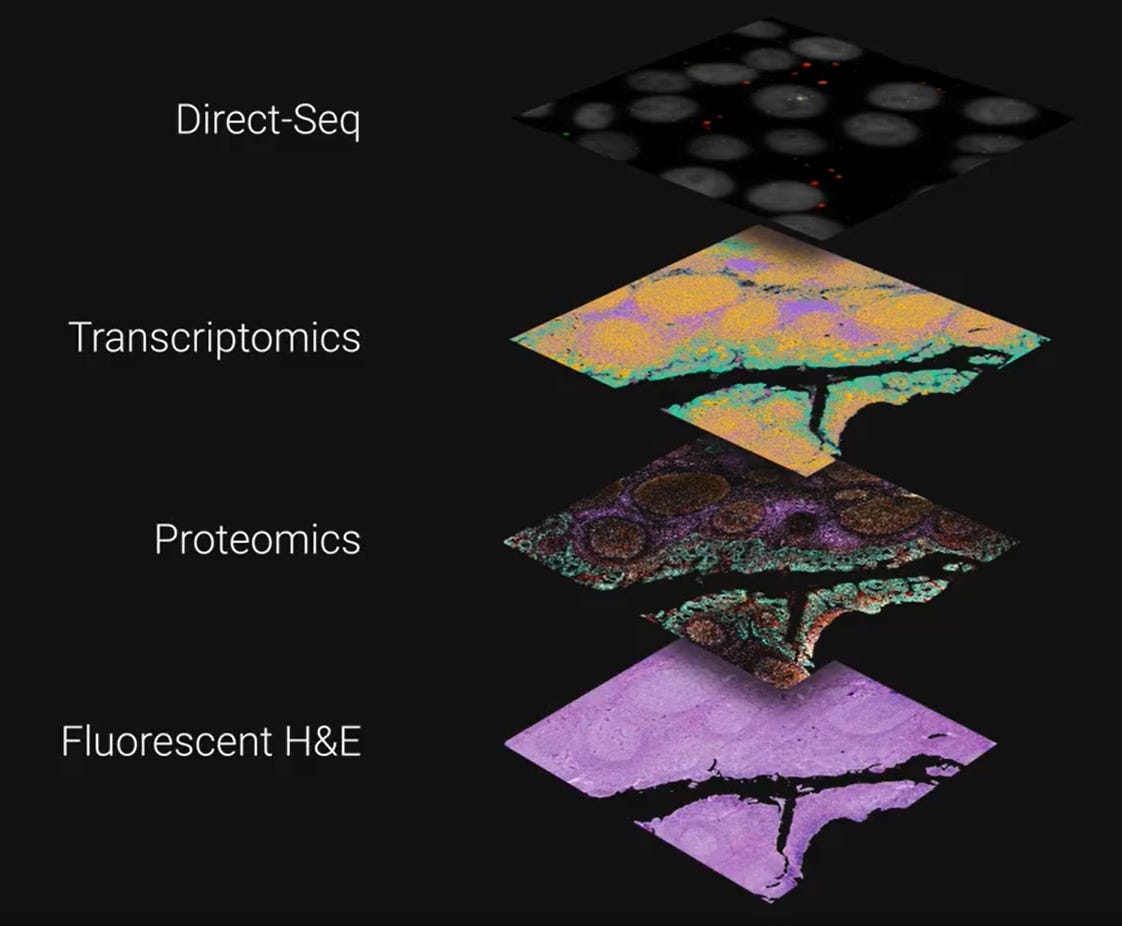
The first method described is DirectSeq, which happens for RNA molecules inside the cells (FFPE?).
They sequence a variable region in the RNA by targetting a known region with a DNA probe thus flanking the region of interest. The RNA sequence is copied into the probe, then amplify it with what looks like Rolling Circle Amplification (RCA).
The region of interest (in blue) is then sequenced on the cell spot where the RCA DNA ball has been produced, just as it would have happened in a cluster in the G4 NGS sequencer, with the proprietary 4-colour chemistry.
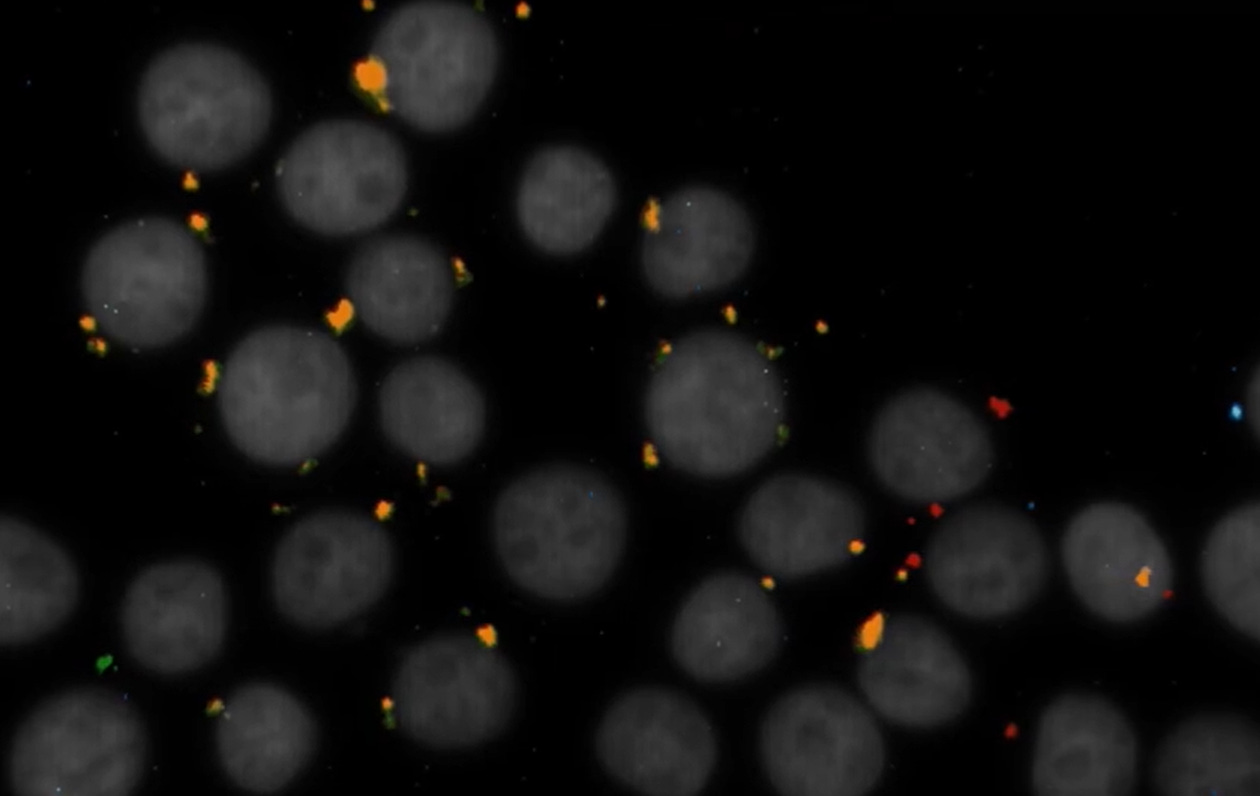
The image of cells with spots changing colours comes with the explanation “we can target the variable region of B-cells and T-cells”. These are cell-specific regions that undergo V(D)J recombination: cutting and pasting letters from alphabet blocks to make unique words. VDJ recombination does this with genes in immune cells, creating billions of "words" like receptors to recognize and fight invaders such as bacteria or viruses.
Another example given is somatic mutations in cancer samples: given a biopsy, one can target the adjacent region of a gene where mutations driving cancer growth are expected, and see where does give a positive mutational hit by reading the sequence that is targetted.
Second mode: in-situ RNA sequencing, they measure gene expression by translating known RNA sequence into DNA via complementary probe and sequence it to identify the RNA transcript. Then the software is used to apply nuclear and cell membrane based segmentation. This method allows for the profiling of hundreds of transcripts simultaneously at single-cell resolution. This feels like it’s the high-resolution spatial equivalent of 10X Genomics Xenium.
Given that everybody is interested in both RNA and proteins when it comes to Spatial Biology offerings, the G4X can also image dozens of proteins in the same tissue section as the RNA profile. The protein imaging part is also part of the cell segmentation image analysis component.
Finally, the technology is able to generate an H&E image using the fluorescent version of the traditional method of staining.
These 4 layers are all applied to the same tissue section.
The G4X looks a lot like the G4, with a footprint similar to other mid-throughput NGS sequencers like the Element Bio AVITI.
G4X Specs
Much larger imaging area than the competitors. Compatible with FFPE.
Specialised tools included, e.g. the transferring of tissue biopsies into the slides. Streamlined 2-day workflow, with 1-day on the instrument, so a total of 3-days TAT.
The comparison below is, I believe, against the 10X Genomics Xenium, with 30x more samples per week:
And at half the cost:
The sequencing of RNA is at up to 70+ base pairs, happens in the instrument.
The G4X is an upgrade on the G4, and existing G4 customers can apply for the upgrade by the end of 2024
These are all the technical details, now I will discuss what this means in terms of the strategic positioning in Spatial Biology and NGS.